RPGeNet is a curated network built from interactions gathered from two interaction databases, BioGRID and STRING, as well as, from the protein interaction text-mining tool, PPaxe.
BioGRID is a genetic and protein interaction database that contains experimentally verified interactions for a plethora of species. STRING is only a protein interaction database that contains both verified interactions and non-verifed predictions. The databases were filterd by species to ensure only human interactions were included in the network (two species hybrid interactions were also discarded). STRING, containing many non-experimentally verified predictions, had all interactions with no evidence filtered out.

STRING database offers a large range of sources of evidences for experimentally validated and predicted protein-to-protein interactions. RPGeNet only includes experimentally validated and no predicted interactions. The graph above shows the proportions of each source found of the experimentally validated interactions used in RPGeNet.

The previous figure shows the distribution of the PPaxe scores given to the interactions detected, the number of PPaxe interactions and the average score depending on the cutoff score chosen. In an attempt to optimize the number interactions while reducing false positive interactions and increasing false-negatives, the cutoff score chosen was 0.65.
To build the complete core network, it was first needed to create the skeleton: a graph network that connects all the driver genes through their shortest paths between them. There are further subgraph levels above the skeleton (level 0 subgraph) that are an extension of that initial graph. The first level is an extension of the skeleton with the addition of parents and children genes that connect to the skeleton but are not themselves within the skeleton. The next level includes parents and children of the genes found within the first level that are not already in the skeleton and level one. The same pattern continues until there is a saturation (no more higher levels can be derived as there are no parent/child edges left to expand the last level), except for the wholegraph defining the complete core network that includes genes that still do not connect with the last network level.

The contribution of evidences from each source to RPGeNet build 2.0.2 is described on the following table:
| SUBGRAPH LEVEL | Skeleton | Level1 | Level2 | Level3 | WholeGraph |
|---|---|---|---|---|---|
| NODES SUMMARY | |||||
| Total UNIQUE NODES | 4 018 | 17 851 | 18 512 | 18 527 | 18 542 |
| Adjacent Nodes | 4 002 | 17 836 | 18 497 | 18 512 | 18 527 |
| Isolated Nodes | 16 | 15 | 15 | 15 | 15 |
| NODES by Source | |||||
| BioGRID | 3 677 | 14 831 | 15 132 | 15 136 | 15 139 |
| STRING | 3 580 | 12 864 | 13 253 | 13 263 | 13 269 |
| PPaxe | 1 407 | 3 016 | 3 054 | 3 056 | 3 062 |
| TOTAL | 8 664 | 30 711 | 31 439 | 31 455 | 31 470 |
| Nodes source "redundancy" | 215.63% | 172.04% | 169.83% | 169.78% | 169.72% |
| EDGES SUMMARY | |||||
| Total DIRECTED EDGES | 35 528 | 932 340 | 1 217 902 | 1 218 017 | 1 218 032 |
| Mutual [A⇌B] | 9 601 | 462 988 | 604 652 | 604 707 | 604 713 |
| Assymetric [A⇀B] | 16 326 | 5 074 | 5 931 | 5 931 | 5 931 |
| Self-loop [A⇀A] | 0 | 1 290 | 2 667 | 2 672 | 2 675 |
| Total Non-Redundant | 25 927 | 469 352 | 613 250 | 613 310 | 613 319 |
| Directed edges "redundancy" | 137.03% | 198.64% | 198.60% | 198.60% | 198.60% |
| EDGES by Source | |||||
| BioGRID all | 22 914 | 518 478 | 623 643 | 623 656 | 623 659 |
| BioGRID only | 21 334 | 483 667 | 579 207 | 579 220 | 579 220 |
| STRING all | 12 563 | 440 111 | 629 167 | 629 265 | 629 271 |
| STRING only | 10 599 | 402 920 | 582 094 | 582 190 | 582 190 |
| PPaxe all | 2 277 | 12 282 | 13 572 | 13 578 | 13 584 |
| PPaxe only | 1 534 | 8 049 | 8 984 | 8 988 | 8 988 |
| TOTAL | 37 754 | 970 871 | 1 266 382 | 1 266 499 | 1 266 514 |
| Edges source "redundancy" | 145.62% | 206.85% | 206.50% | 206.50% | 206.50% |
| EVIDENCE SUMMARY | Skeleton | Level1 | Level2 | Level3 | WholeGraph |
| TOTAL EVIDENCES | 75 447 | 2 371 305 | 3 209 677 | 3 209 856 | 3 209 871 |
| By Class | |||||
| Genetic evidences | 257 | 6 018 | 7 062 | 7 063 | 7 063 |
| Avg. evids x directed edge | 0.007 | 0.006 | 0.006 | 0.006 | 0.006 |
| Physical evidences | 70 561 | 2 342 154 | 3 177 567 | 3 177 739 | 3 177 748 |
| Avg. evids x directed edge | 1.986 | 2.512 | 2.609 | 2.609 | 2.609 |
| Unknown evids (PPaxe) | 4 629 | 23 133 | 25 048 | 25 054 | 25 060 |
| Avg. evids x directed edge | 0.130 | 0.025 | 0.021 | 0.021 | 0.021 |
| By Source | |||||
| BioGRID | 31 726 | 705 485 | 842 798 | 842 815 | 842 818 |
| Physical interactions | 31 469 | 699 467 | 835 736 | 835 752 | 835 755 |
| Genetic Interactions | 257 | 6 018 | 7 062 | 7 063 | 7 063 |
| Avg. evids x directed edge | 0.893 | 0.757 | 0.692 | 0.692 | 0.692 |
| STRING | 39 092 | 1 642 687 | 2 341 831 | 2 341 987 | 2 341 993 |
| Avg. evids x directed edge | 1.100 | 1.762 | 1.923 | 1.923 | 1.923 |
| PPaxe | 4 629 | 23 133 | 25 048 | 25 054 | 25 060 |
| Avg. evids x directed edge | 0.130 | 0.025 | 0.021 | 0.021 | 0.021 |
| EDGES with STRING score | Skeleton | Level1 | Level2 | Level3 | WholeGraph |
| With any STRING score | 12 563 | 440 111 | 629 167 | 629 265 | 629 271 |
| With "experimental" score | 2 696 | 75 075 | 109 191 | 109 231 | 109 235 |
| With "database" score | 9 112 | 371 666 | 541 032 | 541 082 | 541 084 |
| With "text-mining" score | 8 803 | 248 133 | 345 610 | 345 682 | 345 686 |
| With "co-expression" score | 2 227 | 107 246 | 157 974 | 158 008 | 158 010 |
| With "neighborhood" score | 0 | 0 | 0 | 0 | 0 |
| With gene-"fusion" score | 16 | 1 136 | 2 384 | 2 384 | 2 384 |
| With "co-occurence" score | 124 | 6 320 | 8 749 | 8 755 | 8 757 |
A summary of graph statistics for RPGeNet build 2.0.2 at each of the subgraph levels derived from the initial skeleton graph (level 0 subgraph) can be found on the table below:
| SUBGRAPH LEVEL | Skeleton | Level1 | Level2 | Level3 | WholeGraph |
|---|---|---|---|---|---|
| NODES SUMMARY | |||||
| Total #Nodes | 4 018 | 17 851 | 18 512 | 18 527 | 18 542 |
| Isolated Nodes | 16 | 15 | 15 | 15 | 15 |
| Adjacent Driver Genes | 260 of 276 | 261 of 276 | |||
| EDGES SUMMARY | |||||
| Total Directed Edges | 35 528 | 932 340 | 1 217 902 | 1 218 017 | 1 218 032 |
| Mutual [A⇌B] | 9 601 | 462 988 | 604 652 | 604 707 | 604 713 |
| Assymetric [A⇀B] | 16 326 | 5 074 | 5 931 | 5 931 | 5 931 |
| Self-loop [A⇀A] | 0 | 1 290 | 2 667 | 2 672 | 2 675 |
| Total Non-redundant | 25 927 | 469 352 | 613 250 | 613 310 | 613 319 |
| GRAPH STATS | |||||
| Graph Density | 0.0022 | 0.0029 | 0.0036 | 0.0035 | 0.0035 |
| Avg. Clustering Coef. | 0.0613 | 0.1449 | 0.2331 | 0.2331 | 0.2331 |
| Graph Diameter | 8 | 6 | 7 | 8 | 8 |
| Graph Reciprocity | 0.5405 | 0.9946 | 0.9951 | 0.9951 | 0.9951 |
| Avg. Degree | 17.6844 | 104.4580 | 131.5797 | 131.4856 | 131.3809 |
| Avg. Closeness | 0.052 | 0.0529 | 0.0529 | 0.0529 | 0.0295 |
| Betweenness | 10 423.13 | 33 430.86 | 34 988.77 | 35 073.29 | 35 044.91 |
| Avg. Edge Betweenness | 1 629.48 | 980.37 | 812.19 | 814.28 | 814.27 |
| Avg. Coreness | 9.1309 | 54.8258 | 77.0557 | 77.0063 | 76.9456 |
| Avg. Eccentricity | 5.6904 | 4.5666 | 4.9552 | 5.8732 | 5.8691 |
| Avg. Path Length | 3.6155 | 2.8810 | 2.8969 | 2.9000 | 2.9000 |

With an addition of 166 new driver genes now known, RPGeNet was in need of an update. The increase in the number of driver genes partially precipitated an early saturation in the number of levels from the skeleton. Our new network, however, does have less non-redundant interactions and genes in comparison to the previous network. This is mainly due to the extensive filtering of STRING. The previous network included the predictions but the updated network only includes interactions with evidences.
Here we have an example of the result of an initial query for CERKL on RPGeNet.v1 and v2 on screenshots below (top and bottom respectively), to illustrate design changes of Network Explorer web interface but also new functionalities.
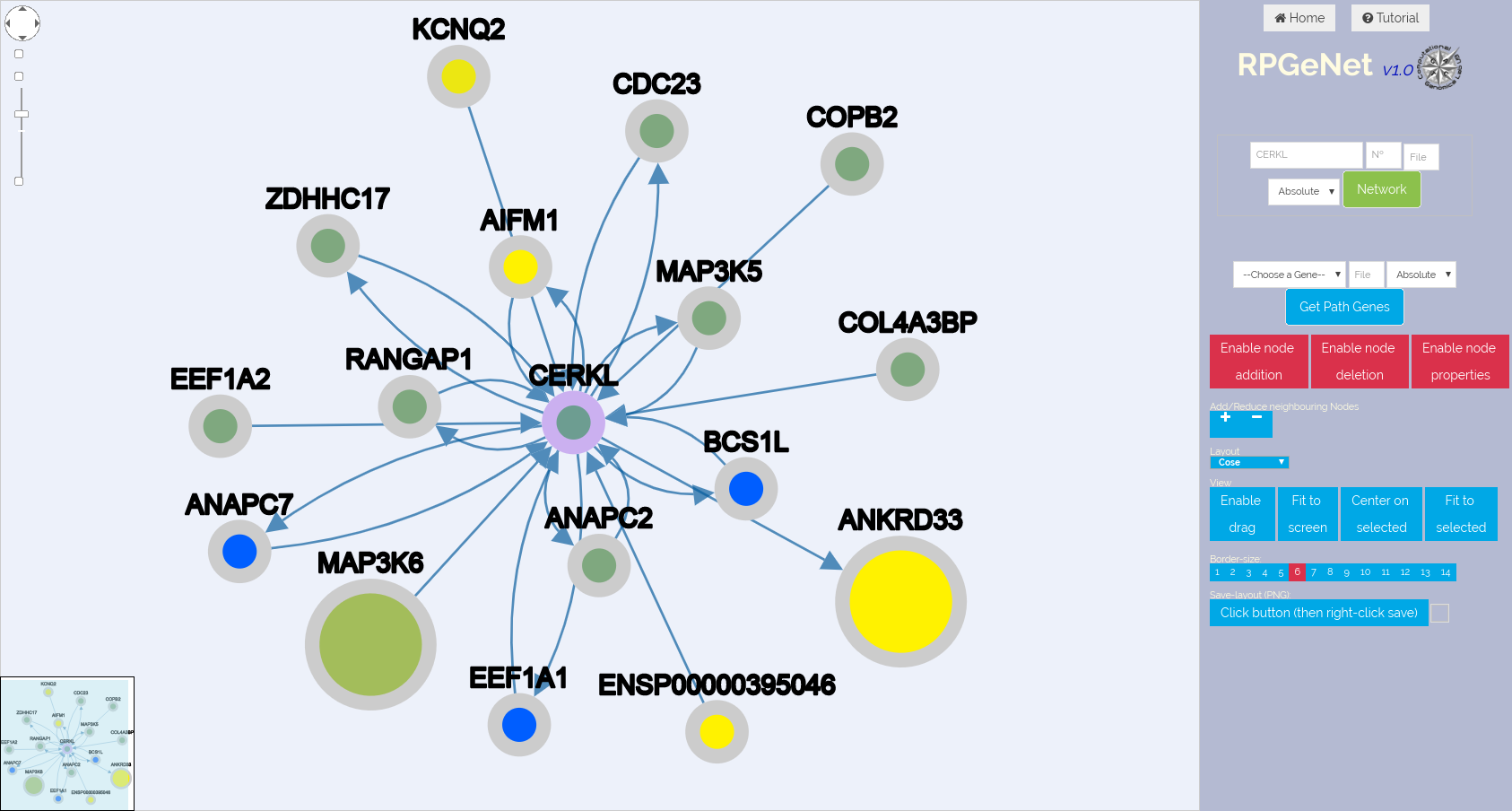

You can download the JSON file containing the saved graph layout to reproduce the CERKL network on RPGeNet.v2 example from the above screenshot.
| VERSION | RPGeNet v1 | RPGeNet v2 | ||
|---|---|---|---|---|
| NODES SUMMARY | Skeleton | WholeGraph | Skeleton | WholeGraph |
| Total #Nodes | 1 294 | 22 372 | 4 018 | 18 542 |
| Isolated Nodes | 7 | 7 | 16 | 15 |
| Adjacent Driver Genes | 103/110 | 103/110 | 260/276 | 261/276 |
| EDGES SUMMARY | Skeleton | WholeGraph | Skeleton | WholeGraph |
| Total Directed Edges | 5 883 | 752 062 | 35 528 | 1 218 032 |
| Mutual [A⇌B] | 1 082 | 319 928 | 9 601 | 604 713 |
| Assymetric [A⇀B] | 3 719 | 106 907 | 16 326 | 5 931 |
| Self-loop [A⇀A] | 0 | 5 299 | 0 | 2 675 |
| Total Non-redundant | 4 801 | 432 134 | 25 927 | 613 319 |
Unfortunately, there are few multi-tissue microarray expression experiments that include retina within their list of tissues. We had to rely on a relatively old microarray experiment (GSE7905) that includes thirty-two different tissues including the retina, liver, brain, skeletal muscle and others. Although the experiment is not recent, the expression values are still useful in finding potentially important pathways associated with retinitis pigmentosa by looking to see if the genes in a pathway all express within the retina. We hope to soon be able to have an updated multitissue expression experiment to use.

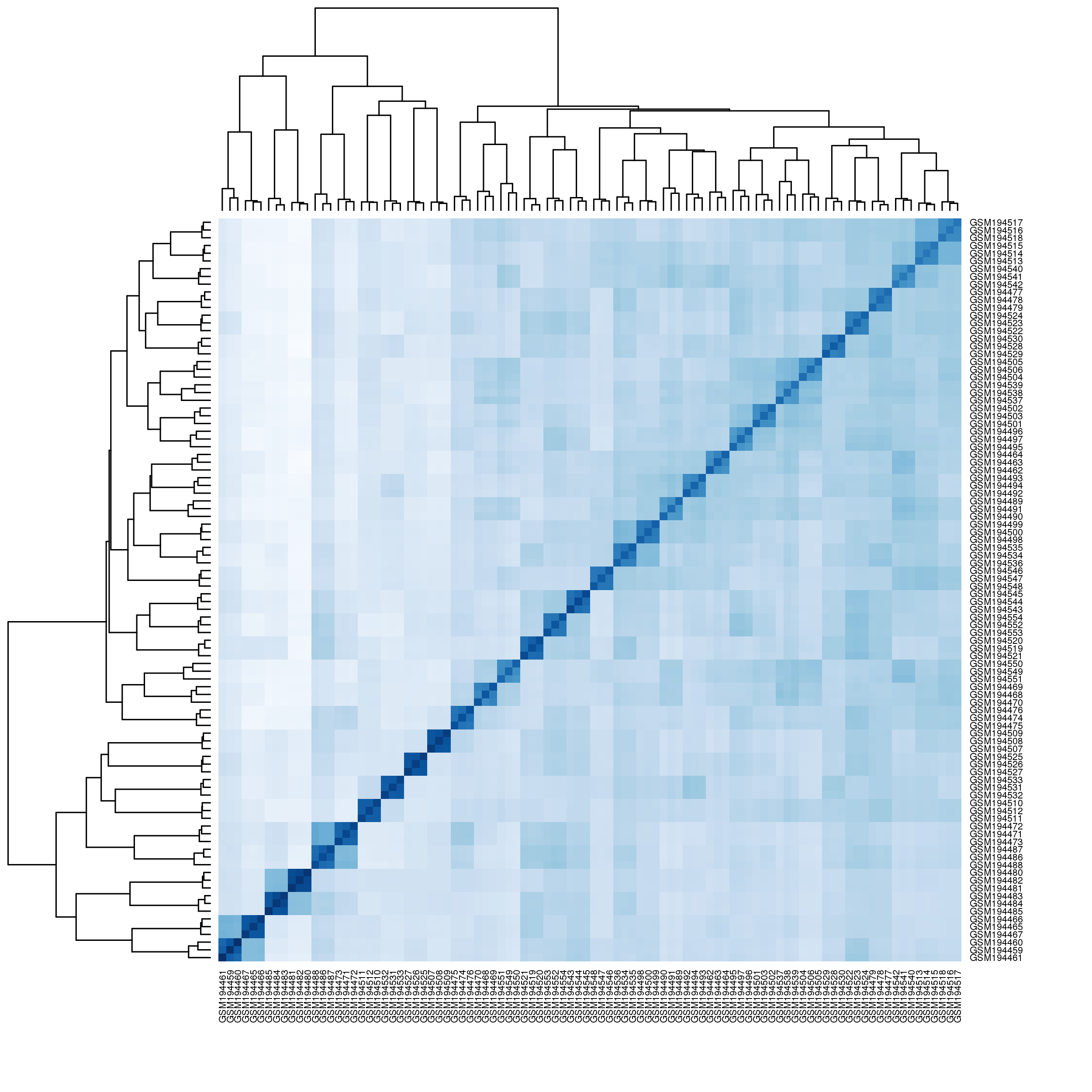
Top twenty relatively overexpressed genes in retina in comparison to all tissues:
| Gene Symbol | Average Expression | t | logFC | P. Value | Adjusted P. Value |
|---|---|---|---|---|---|
| TMEM98 | 1.54276e+01 | 1.19506e+02 | 3.76379e+00 | 1.71574e-78 | 5.64101e-74 |
| UNC119 | 1.52132e+01 | 1.09719e+02 | 4.60785e+00 | 4.54282e-76 | 7.46795e-72 |
| GPX3 | 1.57962e+01 | 9.99257e+01 | 3.96335e+00 | 2.02176e-73 | 2.21571e-69 |
| EFEMP1 | 1.57655e+01 | 8.30937e+01 | 4.06639e+00 | 3.31868e-68 | 2.72779e-64 |
| APOD | 1.54900e+01 | 8.08101e+01 | 4.20815e+00 | 2.02969e-67 | 1.33464e-63 |
| AOC3 | 1.46594e+01 | 7.49936e+01 | 3.50441e+00 | 2.59104e-65 | 1.41980e-61 |
| INPP5K | 1.47864e+01 | 7.08373e+01 | 2.84811e+00 | 1.04379e-63 | 4.90255e-60 |
| SERPINF1 | 1.79001e+01 | 7.02973e+01 | 3.10665e+00 | 1.71338e-63 | 7.04158e-60 |
| SLC22A17 | 1.58003e+01 | 6.94541e+01 | 2.91467e+00 | 3.74310e-63 | 1.36740e-59 |
| SEPT4 | 1.48751e+01 | 6.85776e+01 | 2.78186e+00 | 8.51682e-63 | 2.72644e-59 |
| GJA1 | 1.59067e+01 | 6.85049e+01 | 3.09793e+00 | 9.12187e-63 | 2.72644e-59 |
| FOXC1 | 1.50100e+01 | 6.81441e+01 | 3.80321e+00 | 1.28380e-62 | 3.51741e-59 |
| MGP | 1.71069e+01 | 6.79695e+01 | 2.87181e+00 | 1.51573e-62 | 3.64205e-59 |
| PTP4A3 | 1.45879e+01 | 6.79454e+01 | 2.45700e+00 | 1.55084e-62 | 3.64205e-59 |
| RNASE1 | 1.63907e+01 | 6.58746e+01 | 2.92969e+00 | 1.14808e-61 | 2.51644e-58 |
| CHCHD6 | 1.51307e+01 | 6.55999e+01 | 2.70855e+00 | 1.50417e-61 | 2.90906e-58 |
| C1QTNF5 | 1.29306e+01 | 6.26564e+01 | 4.17481e+00 | 2.91860e-60 | 5.33098e-57 |
| KANK2 | 1.67269e+01 | 6.20026e+01 | 2.52482e+00 | 5.74309e-60 | 9.93797e-57 |
| GPNMB | 1.56794e+01 | 6.17375e+01 | 3.23300e+00 | 7.57265e-60 | 1.24487e-56 |
| ADGRA2 | 1.51704e+01 | 6.12170e+01 | 2.41251e+00 | 1.30741e-59 | 2.04690e-56 |
Top twenty relatively underexpressed genes in retina in comparison to all tissues:
| Gene Symbol | Average Expression | t | logFC | P. Value | Adjusted P. Value |
| SCN8A | 9.21722e+00 | -4.86829e-03 | -1.27044e-03 | 9.96130e-01 | 9.97512e-01 |
|---|---|---|---|---|---|
| KRTAP10_4 | 1.08677e+01 | 4.86202e-03 | 8.24666e-04 | 9.96135e-01 | 9.97512e-01 |
| GLRX2 | 1.25596e+01 | 4.80925e-03 | 7.16169e-04 | 9.96177e-01 | 9.97512e-01 |
| INSL4 | 8.68381e+00 | -4.72755e-03 | -1.61184e-03 | 9.96242e-01 | 9.97521e-01 |
| EHMT1 | 1.11133e+01 | 4.72200e-03 | 1.00922e-03 | 9.96247e-01 | 9.97521e-01 |
| YPEL1 | 1.16304e+01 | 4.29957e-03 | 7.30017e-04 | 9.96582e-01 | 9.97766e-01 |
| RFX8 | 1.03852e+01 | 4.21322e-03 | 1.35426e-03 | 9.96651e-01 | 9.97774e-01 |
| DPF3 | 8.57543e+00 | -3.91878e-03 | -1.14387e-03 | 9.96885e-01 | 9.97948e-01 |
| BANF1 | 1.13208e+01 | -3.85001e-03 | -7.99110e-04 | 9.96940e-01 | 9.97972e-01 |
| PCDHGA6 | 9.22860e+00 | -3.78145e-03 | -1.30935e-03 | 9.96994e-01 | 9.97996e-01 |
| CSNK1G2_AS1 | 9.30784e+00 | -3.46594e-03 | -1.26328e-03 | 9.97245e-01 | 9.98214e-01 |
| WDR62 | 8.99319e+00 | 3.43087e-03 | 9.84490e-04 | 9.97273e-01 | 9.98214e-01 |
| FLJ34790 | 1.07538e+01 | -3.28461e-03 | -9.17140e-04 | 9.97389e-01 | 9.98252e-01 |
| PAM16 | 1.30554e+01 | 2.66893e-03 | 1.43794e-04 | 9.97879e-01 | 9.98577e-01 |
| NCBP2 | 1.30254e+01 | -2.22164e-03 | -1.40418e-04 | 9.98234e-01 | 9.98826e-01 |
| C1ORF127 | 1.00062e+01 | -2.12662e-03 | -8.15441e-04 | 9.98310e-01 | 9.98826e-01 |
| PYY2 | 9.13073e+00 | 1.54440e-03 | 5.05289e-04 | 9.98772e-01 | 9.99168e-01 |
| TEX11 | 9.41583e+00 | 7.98628e-04 | 2.32093e-04 | 9.99365e-01 | 9.99578e-01 |
| C17ORF49 | 1.52699e+01 | 3.14705e-04 | 1.48092e-05 | 9.99750e-01 | 9.99841e-01 |
| RERG | 1.41530e+01 | -8.51990e-05 | -6.10756e-06 | 9.99932e-01 | 9.99934e-01 |